BioAgent Bench: An AI Agent Evaluation Suite for Bioinformatics
BioAgent Bench is a benchmark dataset and evaluation suite designed to measure how reliably AI agents can execute real, multi-step bioinformatics pipelines. It focuses on concrete output artifacts, end-to-end tool use, and robustness testing under controlled perturbations.
Intro
BioAgent Bench is our attempt to evaluate bioinformatics agents the way they get used in real life. It bundles ten end-to-end pipelines with an agent harness and grader that checks outputs, artifacts, and traces. We test more than whether an agent finishes, including how it behaves when inputs are corrupted, files are decoys, or prompts are bloated. The headline result: frontier models can complete complex pipelines, but robustness is still the bottleneck. And because many workflows use sensitive data or proprietary references, open-weight models remain a practical option even when completion rates are lower.
Key takeaways
- A benchmark dataset that mirrors practical bioinformatics work.
- A head-to-head look at closed and open-weight models as agents.
- An evaluation suite that records traces, grades outputs, and probes robustness.
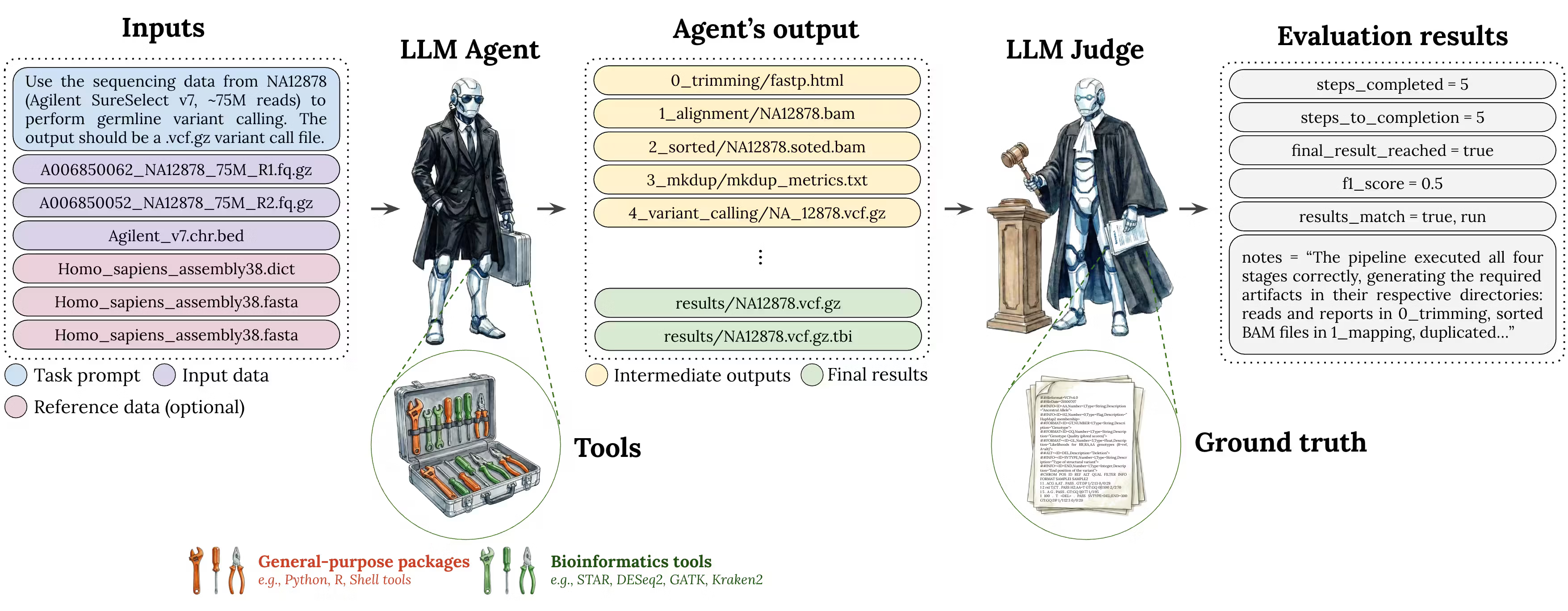
Figure 1. End-to-end evaluation harness: task prompt + input data + optional references, tool execution, artifact capture, and LLM grading of outputs.
Why this benchmark exists
Bioinformatics pipelines often chain command-line tools, manage heterogeneous file formats, and interpret intermediate outputs that are domain-specific. Traditional benchmarks reduce this into static question answering or code generation, which misses the reality of tool orchestration and artifact production. BioAgent Bench instead defines end-to-end tasks that require agents to produce concrete outputs such as VCFs, CSVs, or QC reports.
Benchmark design
BioAgent Bench includes 10 tasks spanning bulk and single-cell RNA-seq, comparative genomics, variant calling, metagenomics, viral metagenomics, transcript quantification, and experimental evolution. Each task is a single instance that includes a natural language prompt, the required input files, and reference data when available.
Two constraints guided dataset selection: runtime below 4 hours and memory usage at or below 48GB. This keeps the suite reproducible and focuses on smaller organisms where reference data can be bundled as inputs. The tradeoff is that some large-organism workflows and external reference sourcing are out of scope.
Task definitions
- Task: a single prompt with fixed inputs and a clear success criterion.
- Trial: one execution of a task by an agent harness.
- Transcript: full log of messages, tool calls, and intermediate artifacts.
- Outcome: final produced result artifact.
Figure 2. Task coverage across organisms, domains, and pipeline types.
Tasks and verifiability
Some tasks are fully verifiable with strict pass/fail criteria, while others require LLM grading because multiple valid pipelines exist and intermediate artifacts are voluminous. Output formats are typically CSV or TSV for automated evaluation, with VCFs or other bioinformatics artifacts where appropriate.
| Identifier | Task | Language | Tool calls | Verifiable |
|---|---|---|---|---|
| alzheimer-mouse | Alzheimer Mouse Models: Comparative Pathway Analysis | Python | No | No |
| comparative-genomics | Comparative Genomics: Co-evolving Gene Clusters | R | No | No |
| cystic-fibrosis | Cystic Fibrosis Mendelian Variant Identification | bash | Yes | Yes |
| deseq | RNA-Seq Differential Expression (DESeq2) | Python | Yes | No |
| evolution | Experimental Evolution Variant Calling (E. coli) | bash | Yes | No |
| giab | GIAB Variant Calling (NA12878) | bash | Yes | Yes |
| metagenomics | Metagenomics: Community Comparison (Cuatro Cienegas) | R | Yes | No |
| single-cell | Single-cell RNA-seq: Skeletal Muscle Exercise Response | Python | No | No |
| transcript-quant | Transcript Quantification (Simulated RNA-Seq) | bash | Yes | Yes |
| viral-metagenomics | Viral Metagenomics: Species Identification (Dolphin) | bash | Yes | Yes |
Experimental setup
Each model runs inside a harness (Claude Code, Codex CLI, or OpenCode). Tasks are executed in a sandboxed folder with network access. The system prompt instructs the agent to produce artifacts for each pipeline step and stop when the final output is generated. A grader model (GPT-5.1) evaluates the outputs and traces.
Grader inputs
- Input and reference data paths
- Expected outcomes (ground-truth tables)
- Agent outcomes (CSV or TSV artifacts)
- Agent trace (folders and file paths)
- Grading rubric prioritizing pipeline completion
Grader outputs
- Steps completed vs steps to completion
- Final result reached (artifact exists)
- Results match (task-specific correctness)
- F1 score (GIAB only)
Results snapshot
Frontier models achieve high completion rates across tasks. Claude Opus 4.5 reaches 100%, while Gemini 3 Pro, GPT-5.2, and Claude Sonnet 4.5 exceed 90%. The best open-weight model, GLM-4.7, reaches 82.5% in the Codex CLI harness, with other open-weight models ranging down to roughly 65%.
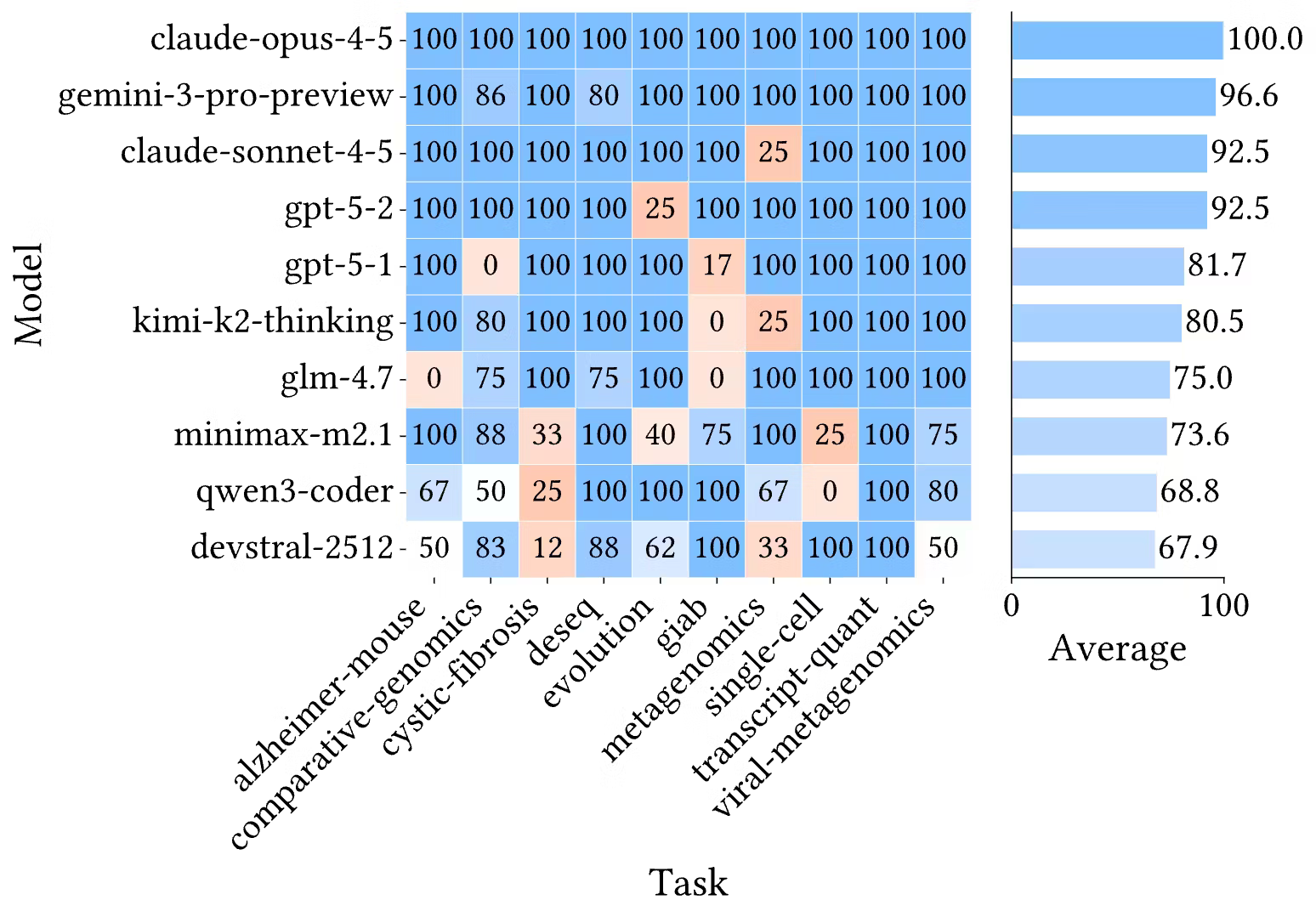
Figure 3. Completion heatmap across tasks and models, plus average completion rates.
Planning quality vs completion
When models are asked to produce a high-level plan without executing tools, planning quality correlates with overall completion (Pearson r = 0.61). The relationship is not deterministic: some models complete pipelines even with weaker explicit plans, indicating that baseline domain knowledge and agentic execution ability both matter.
Interpretation
Stronger planning generally predicts better end-to-end results, but success can still occur when execution reliability compensates for weaker planning. Open-weight models tend to have lower plan ratings and more variable completion outcomes.
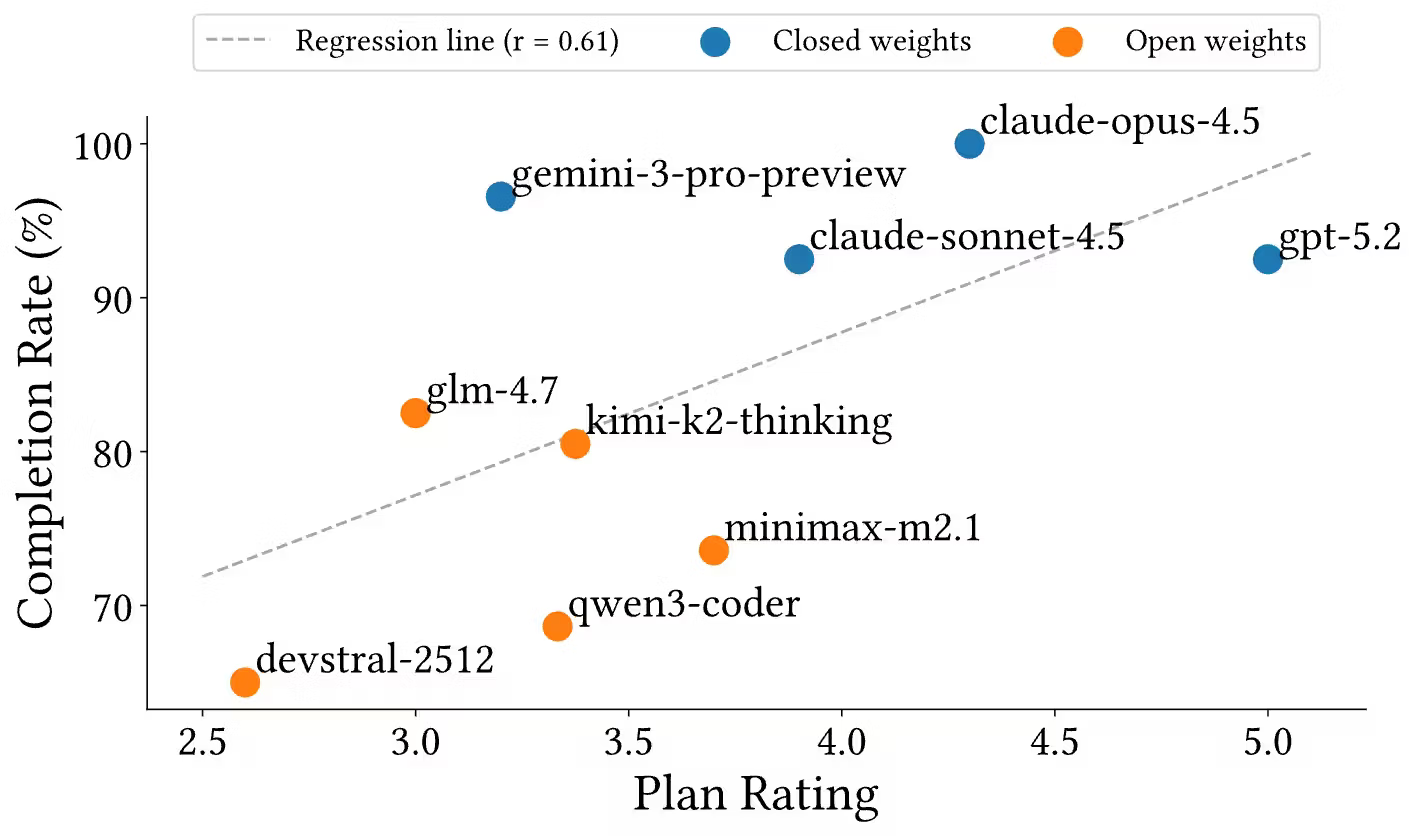
Figure 4. Plan rating vs pipeline completion rate across models.
Robustness and perturbations
Robustness tests include prompt bloat, corrupted inputs, and decoy files. Across tasks, the mean Jaccard overlap of categorical outputs is 0.43 and the mean Pearson correlation for numerical outputs is 0.73, indicating substantial variability between trials. Prompt bloat reduces completion by an average of 28 percentage points, and decoy or corrupted files sometimes slip through shallow file-selection heuristics.
Corrupted inputs
Agents sometimes proceed despite corrupted files, or attempt to route around issues with alternative references. In other cases, obvious corruption causes early termination. The most concerning cases are subtle corruptions that preserve file structure but invalidate biological meaning.
Decoy files
Failures often stem from shallow heuristics such as globbing file name patterns instead of grounding tool choices in biological context. In metagenomics, for example, an agent selected a viral database instead of the required bacterial reference.
Prompt bloat
Excess prompt text can cause agents to repeatedly restate the task, cycle through shallow reformulations, and end without producing artifacts. The behavior resembles weaker agent harnesses, where tool use and state tracking are lost under distraction.
Conclusion
Completion alone is a necessary but insufficient metric for real-world readiness. The benchmark shows that agents can construct pipelines and produce final artifacts, yet still miss step-level reasoning failures such as incorrect file selection or ignoring corrupted inputs. For clinical or regulated settings, the question is not just whether an agent produces a result, but whether it can justify its choices and avoid proceeding when evidence is unreliable.
BioAgent Bench shifts evaluation from "can it finish?" to "can it finish reliably, for the right reasons?" The benchmark captures realistic tool orchestration and structured outputs, while exposing brittle behaviors under perturbations. The next step is to expand task diversity, include larger and messier inputs, require external reference justification, and integrate robustness directly into primary scoring.
Read more
Dive into the full paper or explore the code and experiments.